Publications
- A survey on deep learning in medical image registration: New technologies, uncertainty, evaluation metrics, and beyond
- Authors: Junyu Chen*, Yihao Liu*, Shuwen Wei*, Zhangxing Bian, Shalini Subramanian, Aaron Carass, Jerry L. Prince, Yong Du
- Published in: Medical Image Analysis
- Paper link: https://www.sciencedirect.com/science/article/pii/S1361841524003104
- PDF:

- Code: MedImgReg_Survey
- Description: Deep learning technologies have dramatically reshaped the field of medical image registration over the past decade. The initial developments, such as regression-based and U-Net-based networks, established the foundation for deep learning in image registration. Subsequent progress has been made in various aspects of deep learning-based registration, including similarity measures, deformation regularizations, network architectures, and uncertainty estimation. These advancements have not only enriched the field of image registration but have also facilitated its application in a wide range of tasks, including atlas construction, multi-atlas segmentation, motion estimation, and 2D-3D registration. In this paper, we present a comprehensive overview of the most recent advancements in deep learning-based image registration. We begin with a concise introduction to the core concepts of deep learning-based image registration. Then, we delve into innovative network architectures, loss functions specific to registration, and methods for estimating registration uncertainty. Additionally, this paper explores appropriate evaluation metrics for assessing the performance of deep learning models in registration tasks. Finally, we highlight the practical applications of these novel techniques in medical imaging and discuss the future prospects of deep learning-based image registration.
- Vector Field Attention for Deformable Image Registration
- Authors: Yihao Liu, Junyu Chen, Lianrui Zuo, Aaron Carass, Jerry L. Prince
- Published in: Journal of Medical Imaging
- Paper link: SPIE Digital Library
- PDF:

- Code: VFA
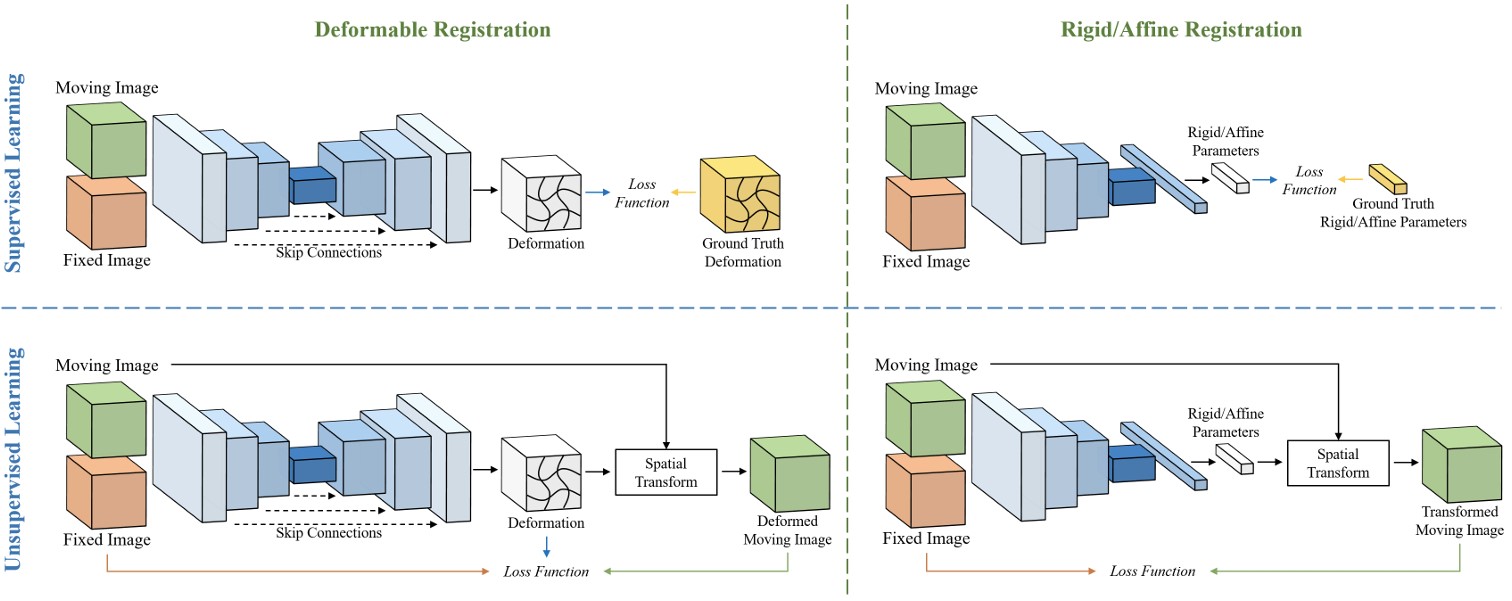
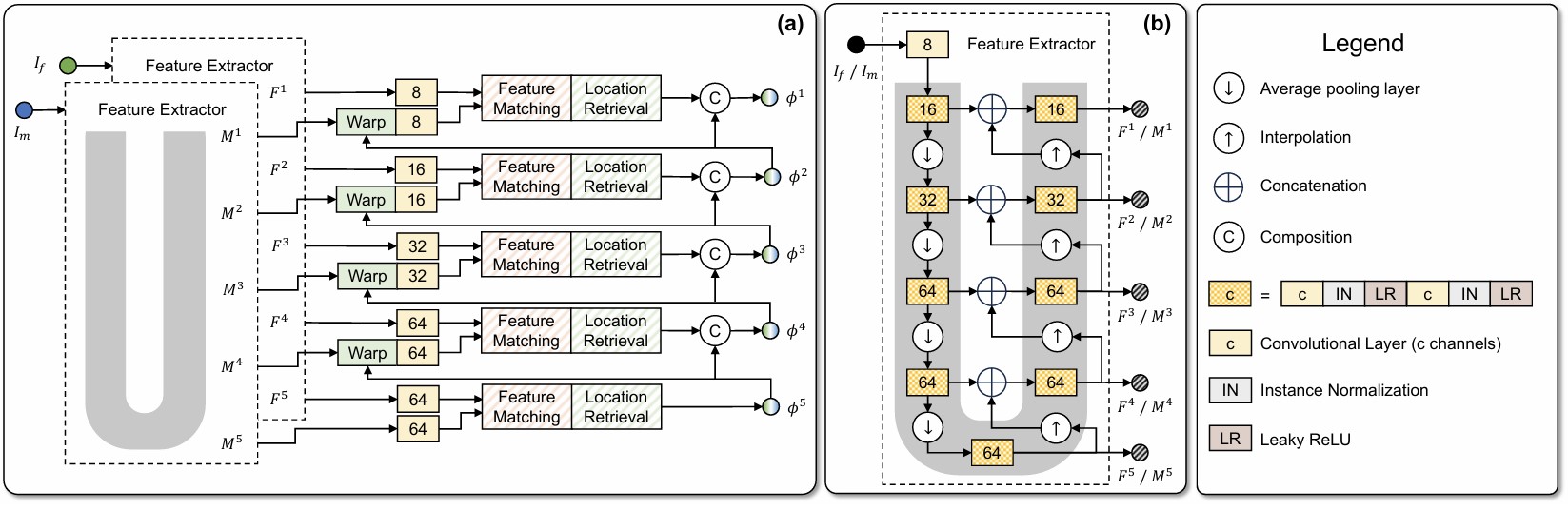
- Description: Deformable image registration establishes non-linear spatial correspondences between fixed and moving images. Deep learning-based deformable registration methods have been widely studied in recent years due to their speed advantage over traditional algorithms as well as their better accuracy. Most existing deep learning-based methods require neural networks to encode location information in their feature maps and predict displacement or deformation fields though convolutional or fully connected layers from these high-dimensional feature maps. In this work, we present Vector Field Attention (VFA), a novel framework that enhances the efficiency of the existing network design by enabling direct retrieval of location correspondences. VFA uses neural networks to extract multi-resolution feature maps from the fixed and moving images and then retrieves pixel-level correspondences based on feature similarity. The retrieval is achieved with a novel attention module without the need of learnable parameters. VFA is trained end-to-end in either a supervised or unsupervised manner. We evaluated VFA for intra- and inter-modality registration and for unsupervised and semi-supervised registration using public datasets, and we also evaluated it on the Learn2Reg challenge. Experimental results demonstrate the superior performance of VFA compared to existing methods.
- On Finite Difference Jacobian Computation in Deformable Image Registration
- Authors: Yihao Liu, Junyu Chen, Shuwen Wei, Aaron Carass, and Jerry Prince
- Published in: International Journal of Computer Vision
- Paper link: https://link.springer.com/article/10.1007/s11263-024-02047-1
- PDF:

- Code: digital_diffeomorphism
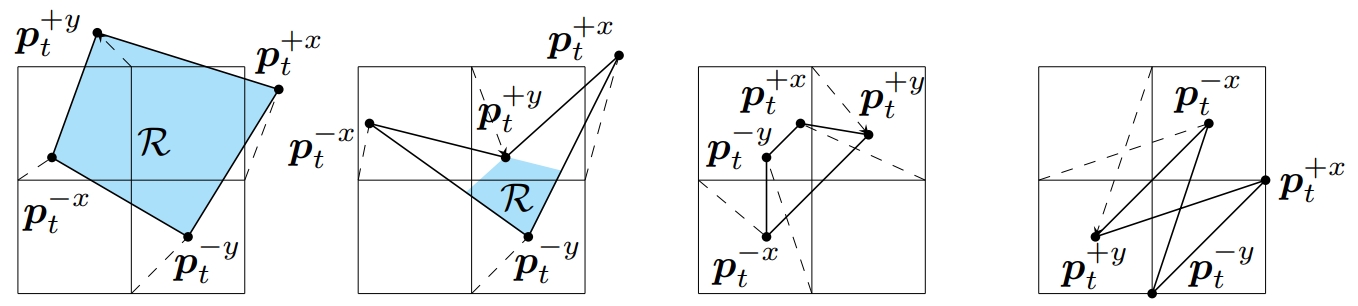
- Description: Producing spatial transformations that are diffeomorphic has been a central problem in deformable image registration. As a diffeomorphic transformation should have positive Jacobian determinant |J| everywhere, the number of voxels with |J|<0 has been used to test for diffeomorphism and also to measure the irregularity of the transformation. For digital transformations, |J| is commonly approximated using central difference, but this strategy can yield positive |J|’s for transformations that are clearly not diffeomorphic—even at the voxel resolution level. To show this, we first investigate the geometric meaning of different finite difference approximations of |J|. We show that to determine diffeomorphism for digital images, use of any individual finite difference approximations of |J| is insufficient. We show that for a 2D transformation, four unique finite difference approximations of |J|’s must be positive to ensure the entire domain is invertible and free of folding at the pixel level. We also show that in 3D, ten unique finite differences approximations of |J|’s are required to be positive. Our proposed digital diffeomorphism criteria solves several errors inherent in the central difference approximation of |J| and accurately detects non-diffeomorphic digital transformations.
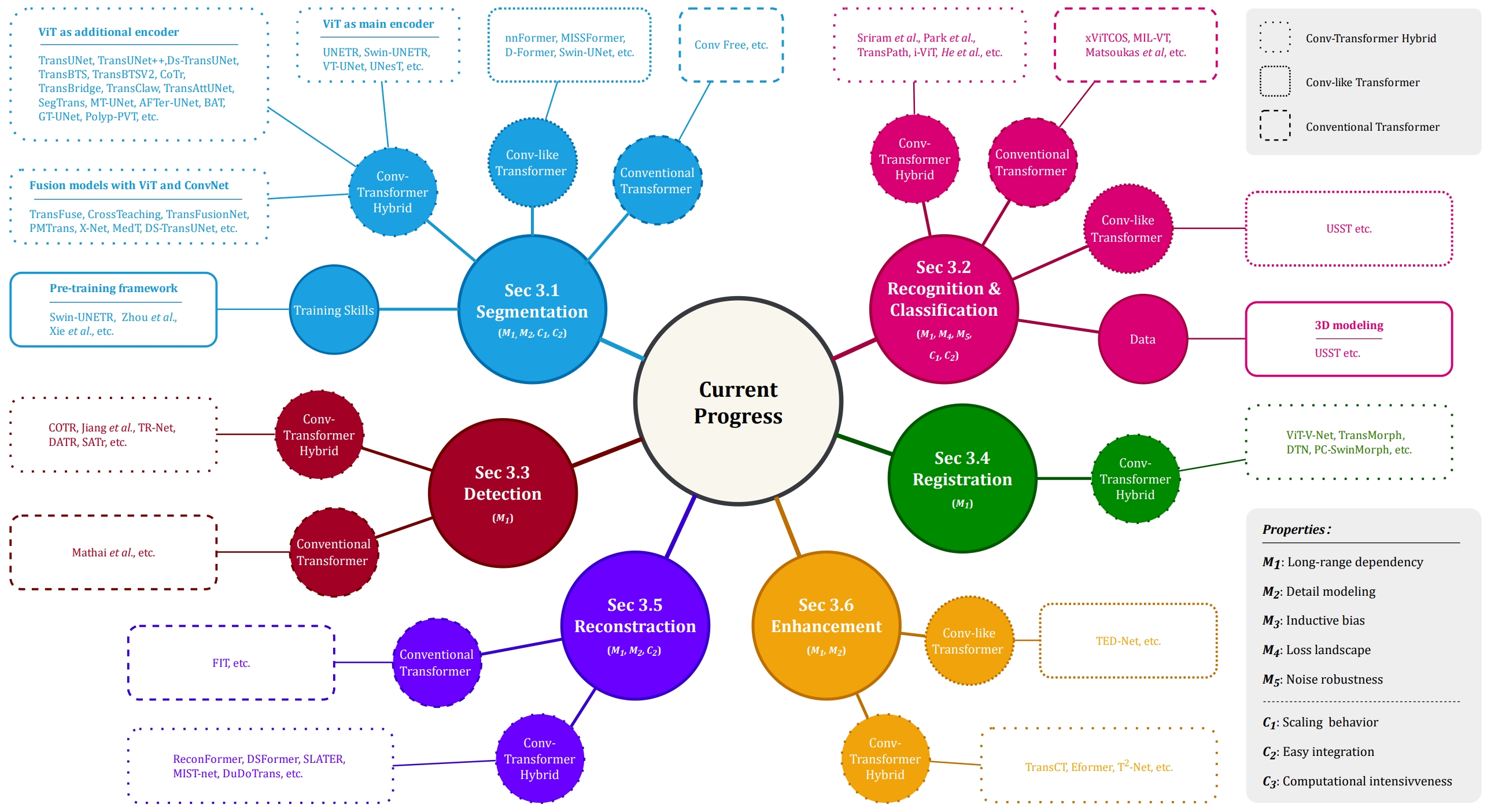
- Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspectives.
- Authors: Jun Li*, Junyu Chen*, Yucheng Tang*, Ce Wang, Bennett A. Landman, and S. Kevin Zhou (*: Equal Contribution)
- Published in: Medical Image Analysis
- Paper link: https://www.sciencedirect.com/science/article/abs/pii/S1361841523000233
- PDF:

- Description: Transformer, one of the latest technological advances of deep learning, has gained prevalence in natural language processing or computer vision. Since medical imaging bear some resemblance to computer vision, it is natural to inquire about the status quo of Transformers in medical imaging and ask the question: can the Transformer models transform medical imaging? In this paper, we attempt to make a response to the inquiry. After a brief introduction of the fundamentals of Transformers, especially in comparison with convolutional neural networks (CNNs), and highlighting key defining properties that characterize the Transformers, we offer a comprehensive review of the state-of-the-art Transformer-based approaches for medical imaging and exhibit current research progresses made in the areas of medical image segmentation, recognition, detection, registration, reconstruction, enhancement, etc.
- Unsupervised Learning of Diffeomorphic Image Registration via TransMorph
- Authors: Junyu Chen, Eric C. Frey, and Yong Du
- Published in: International Workshop on Biomedical Image Registration (WBIR), 2022 (Long oral presentation)
- Paper link: https://openreview.net/forum?id=uwIo__2xnTO
- PDF:

- Code: TransMorph-TVF
- Description: In this work, we propose a learning-based framework for unsupervised and end-to-end learning of diffeomorphic image registration. Specifically, the proposed network learns to produce and integrate timedependent velocity fields in an LDDMM setting. The proposed method guarantees a diffeomorphic transformation and allows the transformation to be easily and accurately inverted. We also showed that, without explicitly imposing a diffeomorphism, the proposed network can provide a significant performance gain while preserving the spatial smoothness in the deformation. The proposed method outperforms the state-of-theart registration methods on two widely used publicly available datasets, indicating its effectiveness for image registration.

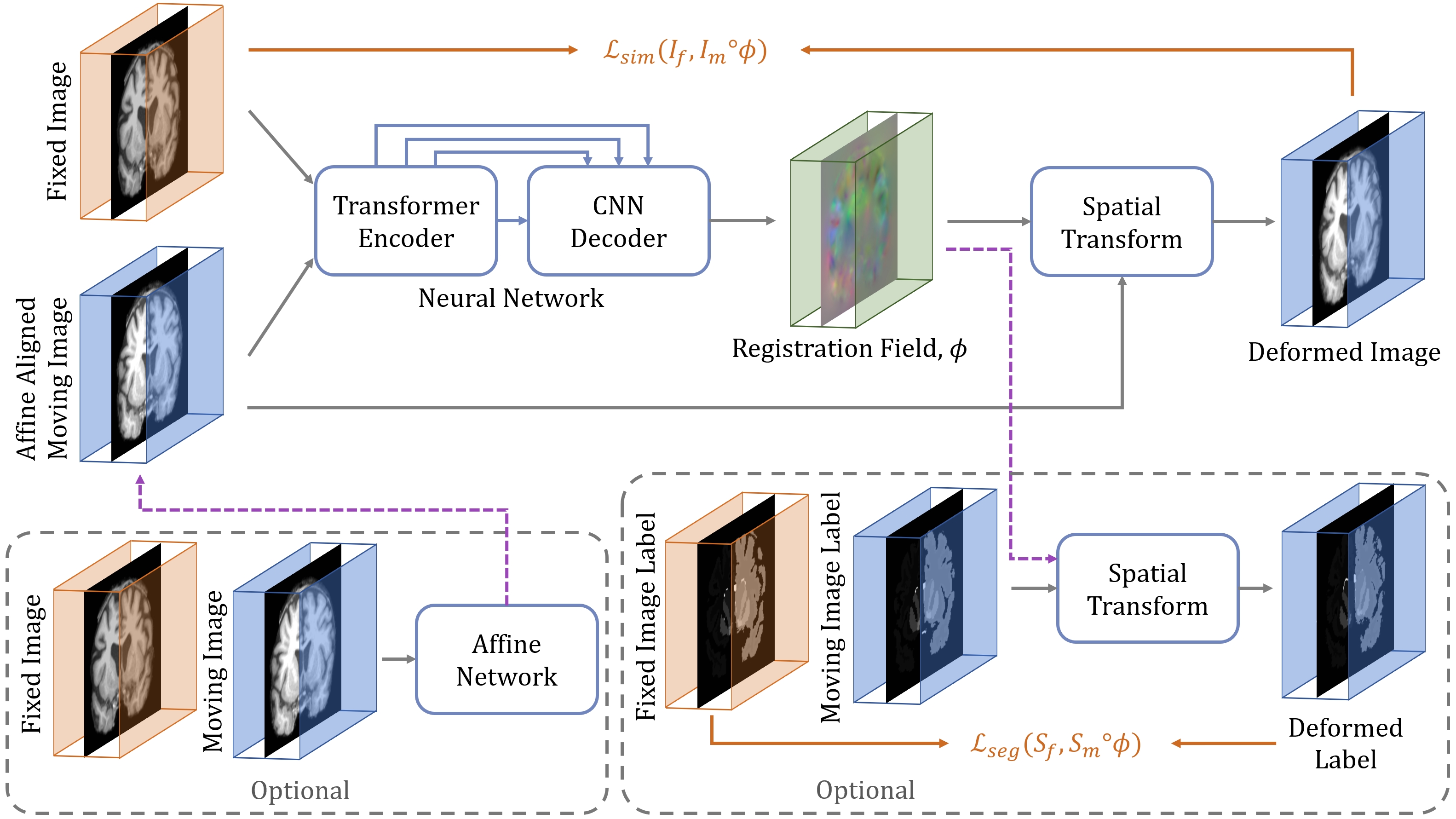
- TransMorph: Transformer for Unsupervised Medical Image Registration
- Authors: Junyu Chen, Yong Du, Yufan He, William P. Segars, Ye Li, and Eric C. Frey
- Published in: Medical Image Analysis
- Paper link: https://www.sciencedirect.com/science/article/pii/S1361841522002432
- PDF:

- Code: TransMorph
- Description: Transformers may be a strong candidate for image registration because their self-attention mechanism enables a more precise comprehension of the spatial correspondence between moving and fixed images. In this paper, we present TransMorph, a hybrid Transformer-ConvNet model for volumetric medical image registration. We also introduce three variants of TransMorph, with two diffeomorphic variants ensuring the topology-preserving deformations and a Bayesian variant producing a well-calibrated registration uncertainty estimate.
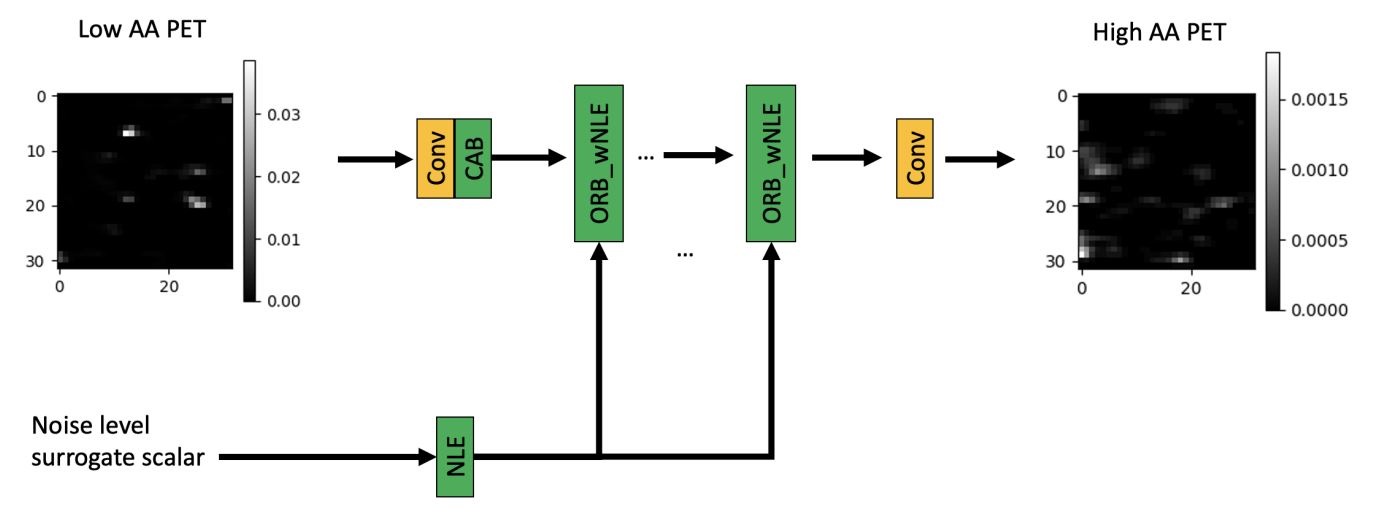
- A Noise-level-aware Framework for PET Image Denoising
- Authors: Ye Li, Jianan Cui, Junyu Chen, Guodong Zeng, Scott Wollenweber, Floris Jansen, Se-In Jang, Kyungsang Kim, Kuang Gong, Quanzheng Li
- Paper link: https://arxiv.org/abs/2203.08034
- PDF:

- Description: In PET, the amount of relative (signal-dependent) noise present in different body regions can be significantly different and is inherently related to the number of counts present in that region. The number of counts in a region depends, in principle and among other factors, on the total administered activity, scanner sensitivity, image acquisition duration, radiopharmaceutical tracer uptake in the region, and patient local body morphometry surrounding the region. In theory, less amount of denoising operations is needed to denoise a high-count (low relative noise) image than images a low-count (high relative noise) image, and vice versa. The current deep-learning-based methods for PET image denoising are predominantly trained on image appearance only and have no special treatment for images of different noise levels. Our hypothesis is that by explicitly providing the local relative noise level of the input image to a deep convolutional neural network (DCNN), the DCNN can outperform itself trained on image appearance only. To this end, we propose a noise-level-aware framework denoising framework that allows embedding of local noise level into a DCNN.
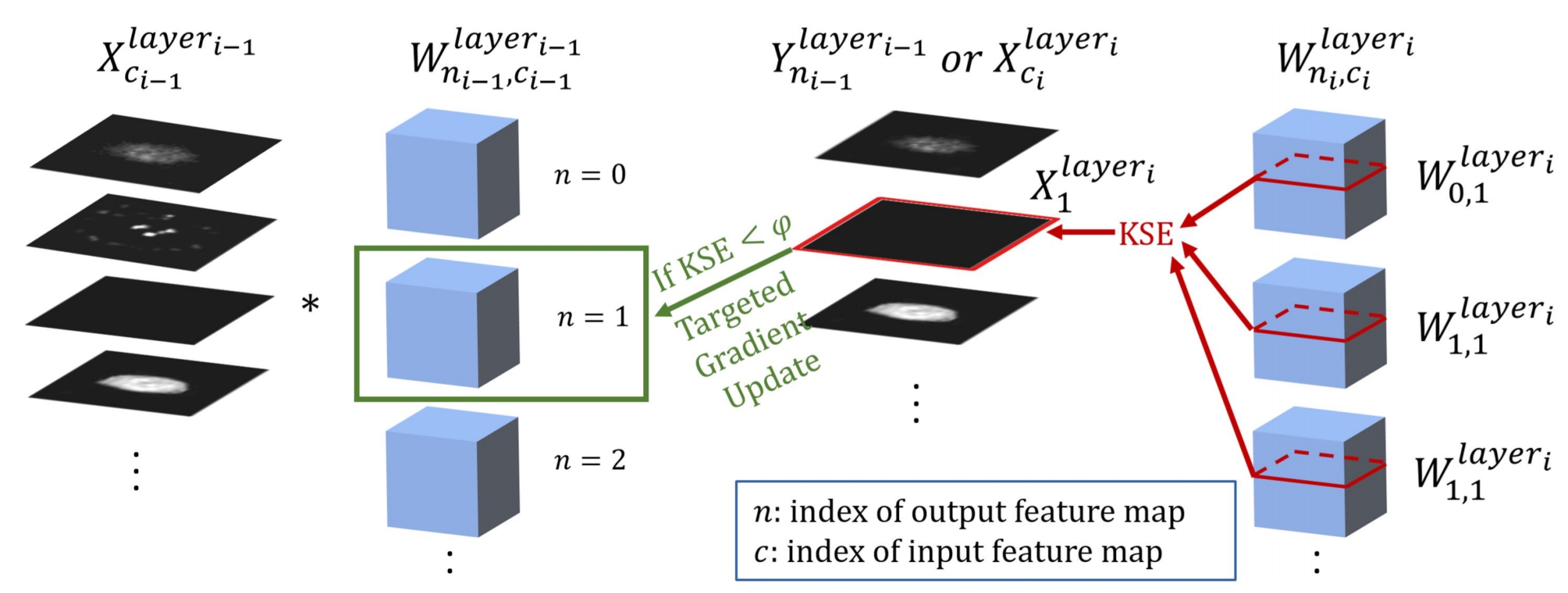
- Targeted Gradient Descent: A Novel Method for Convolutional Neural Networks Fine-tuning and Online-learning
- Authors: Junyu Chen, Evren Asma, and Chung Chan
- Published in: MICCAI, 2021 (Oral presentation)(provisionally accepted, top 13% of 1630 papers)
- Paper link: https://link.springer.com/chapter/10.1007/978-3-030-87199-4_3
- PDF:

- Supplementary file:

- Code: Patent processing. Not available.
- Description: A ConvNet is usually trained and then tested using images drawn from the same distribution. To generalize a ConvNet to various tasks often requires a complete training dataset that consists of images drawn from different tasks. In most scenarios, it is nearly impossible to collect every possible representative dataset as a priori. The new data may only become available after the ConvNet is deployed in clinical practice. ConvNet, however, may generate artifacts on out-of-distribution testing samples. In this study, we present Targeted Gradient Descent, a novel fine-tuning method that can extend a pre-trained network to a new task without revisiting data from the previous task while preserving the knowledge acquired from previous training. To a further extent, the proposed method also enables online learning of patient-specific data to enhance the network’s generalization capability in real-world applications.
- Learning Fuzzy Clustering for SPECT/CT Segmentation via Convolutional Neural Networks
- Authors: Junyu Chen, Ye Li, Licia P. Luna, Hyun Woo Chung, Steven P. Rowe, Yong Du, Lilja B. Solnes, and Eric C. Frey
- Published in: Medical Physics
- Paper link: https://aapm.onlinelibrary.wiley.com/doi/10.1002/mp.14903
- PDF:

- Code: Semi-supervised_FCM_Loss_for_Segmentation
- Description: We present a new unsupervised segmentation loss function and its semi- and supervised variants for training a convolutional neural network (ConvNet). The loss functions were developed based on the objective function of the classical Fuzzy C-means (FCM) algorithm. The first proposed loss function can be computed within the input image itself without any ground truth labels, and is thus unsupervised; the proposed supervised loss function follows the traditional paradigm of the deep learning-based segmentation methods and leverages ground truth labels during training. The last loss function is a combination of the first and the second and includes a weighting parameter, which enables semi-supervised segmentation using deep learning neural network.
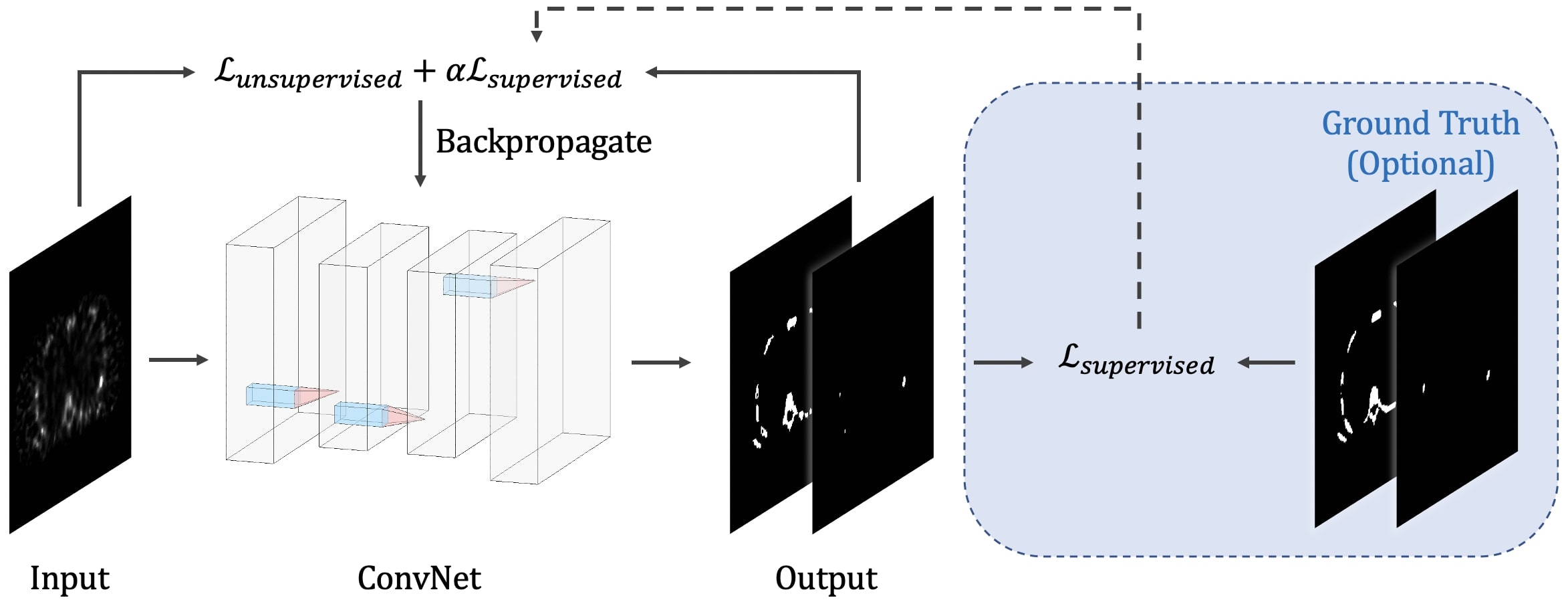
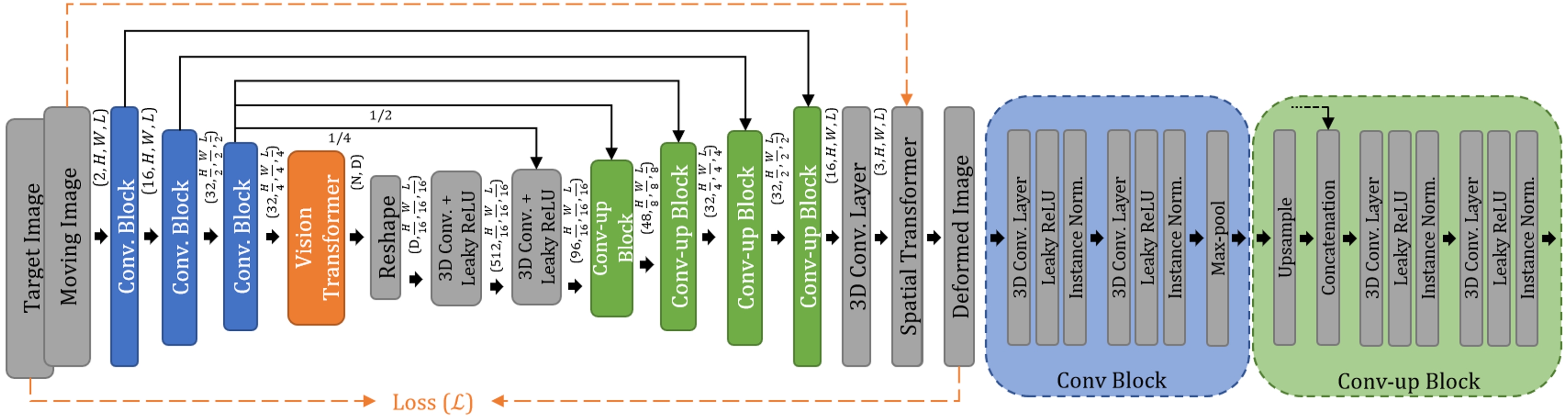
- ViT-V-Net: Vision Transformer for Unsupervised Volumetric Medical Image Registration
- Authors: Junyu Chen, Yufan He, Eric C. Frey, Ye Li, Yong Du
- Published in: Medical Imaging with Deep Learning, 2021
- Paper link: https://arxiv.org/abs/2104.06468
- PDF:

- Code: ViT-V-Net_for_3D_Image_Registration
- Description: We present ViT-V-Net that bridges Vision Transformer and V-Net for volumetric medical image registration. The experimental results demonstrate that the proposed architecture achieves superior performance to several state-of-the-art registration methods.
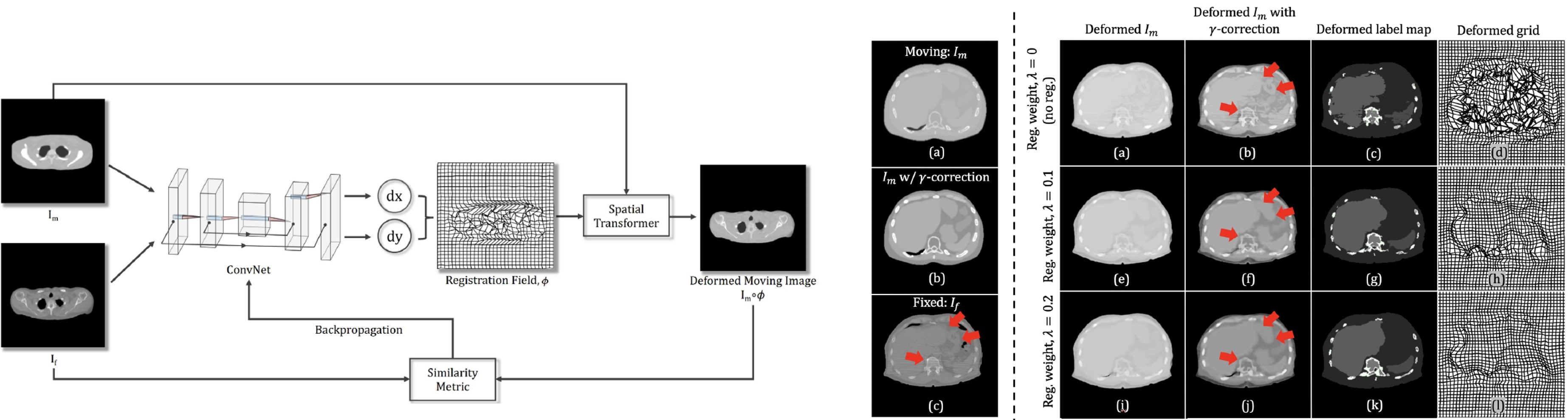
- Generating anthropomorphic phantoms using fully unsupervised deformable image registration with convolutional neural networks
- Authors: Junyu Chen, Ye Li, Yong Du, Eric C. Frey
- Published in: Medical Physics, Dec. 2020 (Editor’s Choice)
- Paper link: https://aapm.onlinelibrary.wiley.com/doi/abs/10.1002/mp.14545
- PDF:

- Code: Fully_Unsupervised_CNN_Registration_Keras
- Description: We treat CNN as an optimization tool that iteratively minimizes the loss function via reparametrization in each iteration. This means that the algorithm is fully unsupervised and thus no prior training is required. We generate phantom variations by warpping an XCAT phantom to capture the anatomical variations within the real human CT images.
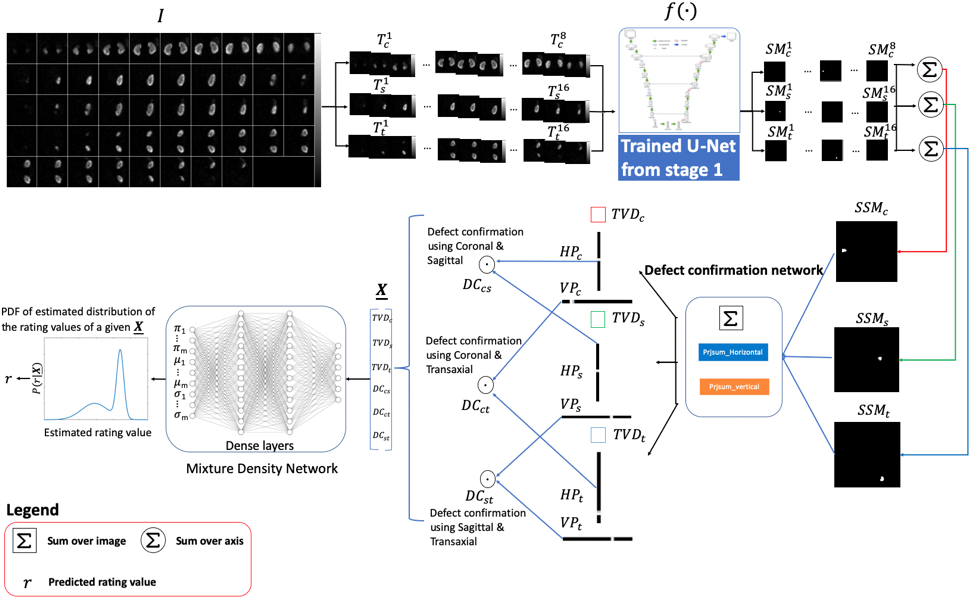
- DeepAMO: A Multi-Slice, Multi-View Anthropomorphic Model Observer for Visual Detection Tasks Performed on Volume Images
- Authors: Ye Li, Junyu Chen, Justin L. Brown, S. Ted Treves, Xinhua Cao, Frederic H. Fahey, George Sgouros, Wesley E. Bolch, Eric C. Frey
- Published in: Journal of Medical Imaging’s special section: Perspectives in Human and Model Observer Performance, 2020
- Paper link: SPIE. Digital Library
- PDF:

- Code: From GaryLi’s site
- Description: We have developed a deep learning-based anthropomorphic model observer (DeepAMO) for image quality evaluation of multi-orientation, multi-slice image sets with respect to a clinically realistic 3D defect detection task. The input to the DeepAMO is a composite image, typical of that used to view 3D volumes in clinical practice. The output is a rating value designed to mimic human observer’s defect detection performance. The main contributions of this paper are threefold. First, we propose a hypothetical model of the decision process of a reader performing a detection task using a 3D volume. Second, we propose a projection-based defect confirmation network architecture to confirm defect present in two different slicing orientations. Third, we propose a novel calibration method that ‘learns’ the underlying distribution of observer ratings from the human observer rating data (thus modeling inter- or intra- observer variability) using a Mixture Density Network.
- Medical Image Segmentation via Unsupervised Convolutional Neural Network
- Authors: Junyu Chen, Eric C. Frey
- Published in: Medical Imaging with Deep Learning, 2020
- Paper link: https://openreview.net/forum?id=XrbnSCv4LU
- PDF:

- Code: Unsuprevised_Seg_via_CNN
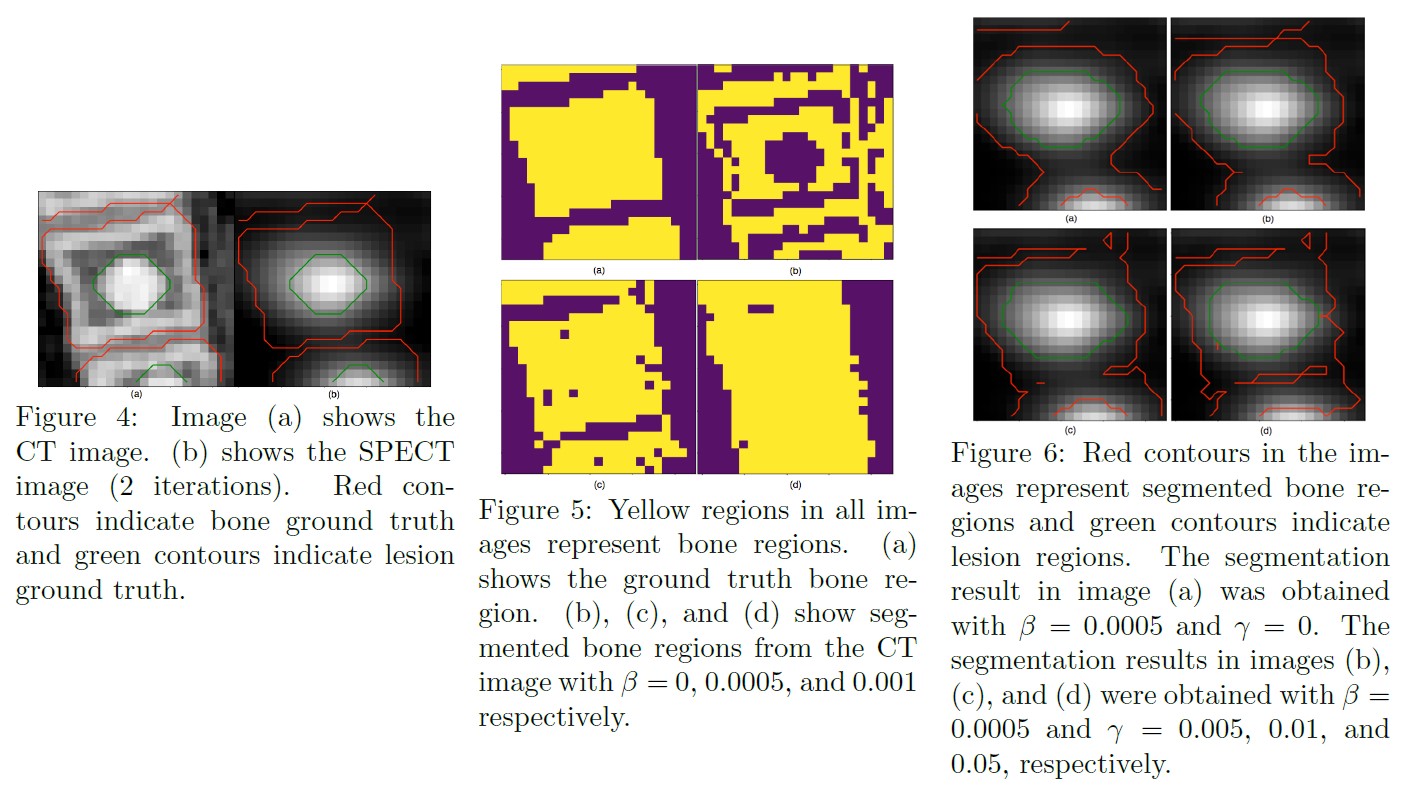
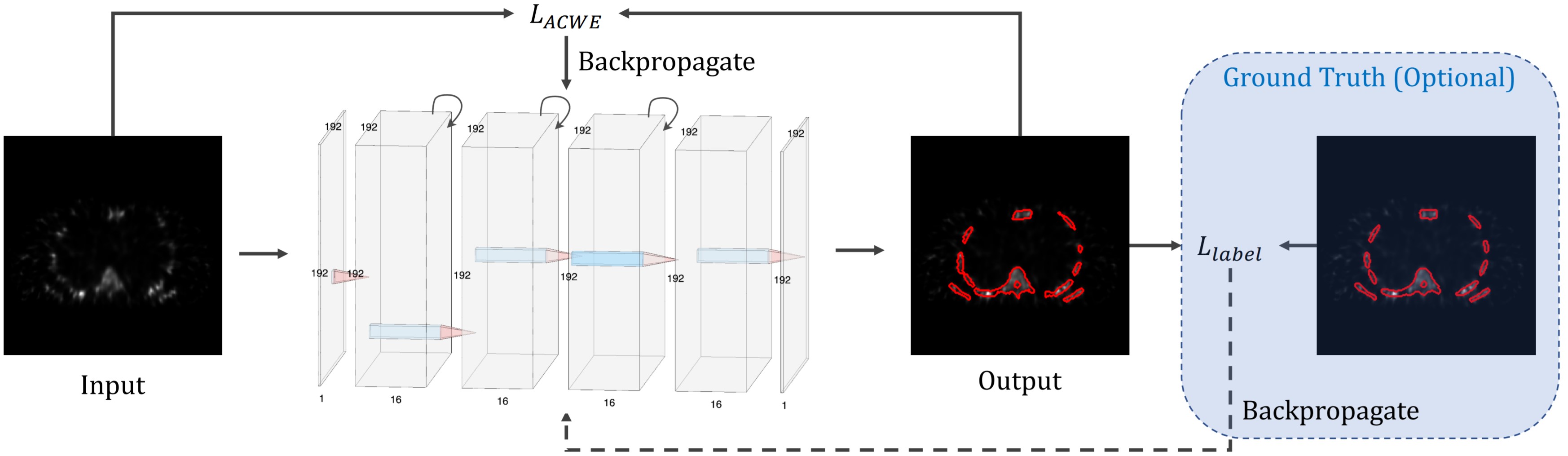
- Description: For the majority of the learning-based segmentation methods, a large quantity of highquality training data is required. In this paper, we present a novel learning-based segmentation model that could be trained semi- or un- supervised. Specifically, in the unsupervised setting, we parameterize the Active contour without edges (ACWE) framework via a convolutional neural network (ConvNet), and optimize the parameters of the ConvNet using a self-supervised method. In another setting (semi-supervised), the auxiliary segmentation ground truth is used during training. We show that the method provides fast and high-quality bone segmentation in the context of single-photon emission computed tomography (SPECT) image.
- Incorporating CT prior information in the robust fuzzy C-means algorithm for QSPECT image segmentation
- Authors: Junyu Chen, Abhinav K Jha, Eric C. Frey
- Published in: Medical Imaging 2019: Image Processing
- Paper link: SPIE Digital Library
- PDF:

- Code: SPECT-CT-Seg-RFCM
- Description: We propose and evaluate a fuzzy C-means (FCM) clustering based semi-automatic segmentation method on quantitative Tc-99m MDP quantitative SPECT/CT. We propose to incorporate information from registered CT images, which can be used to segment normal bones quite readily, into the FCM segmentation algorithm. The proposed method modifies the objective function of the robust fuzzy C-means (RFCM) method to include prior information about bone from CT images and spatial information from the SPECT image to allow for simultaneously segmenting lesion and bone in BQSPECT/CT images.